While a postdoctoral associate in Pratyush Tiwary's lab at the University of Maryland, my research mainly focused on two fairly disparate topics: developing a rough version of energy-entropy disentanglement in a low-dimensional latent space discovered through a generative machine learning technique called the state predictive information bottleneck (SPIB) and using this SPIB method to find good reaction coordinates (RCs) for the nucleation of organic molecules from aqueous solution. To our knowledge, the biased simulations analyzed in this work are the first to sample nucleation of glycine monomers from solution without the use of seeded MD approaches.

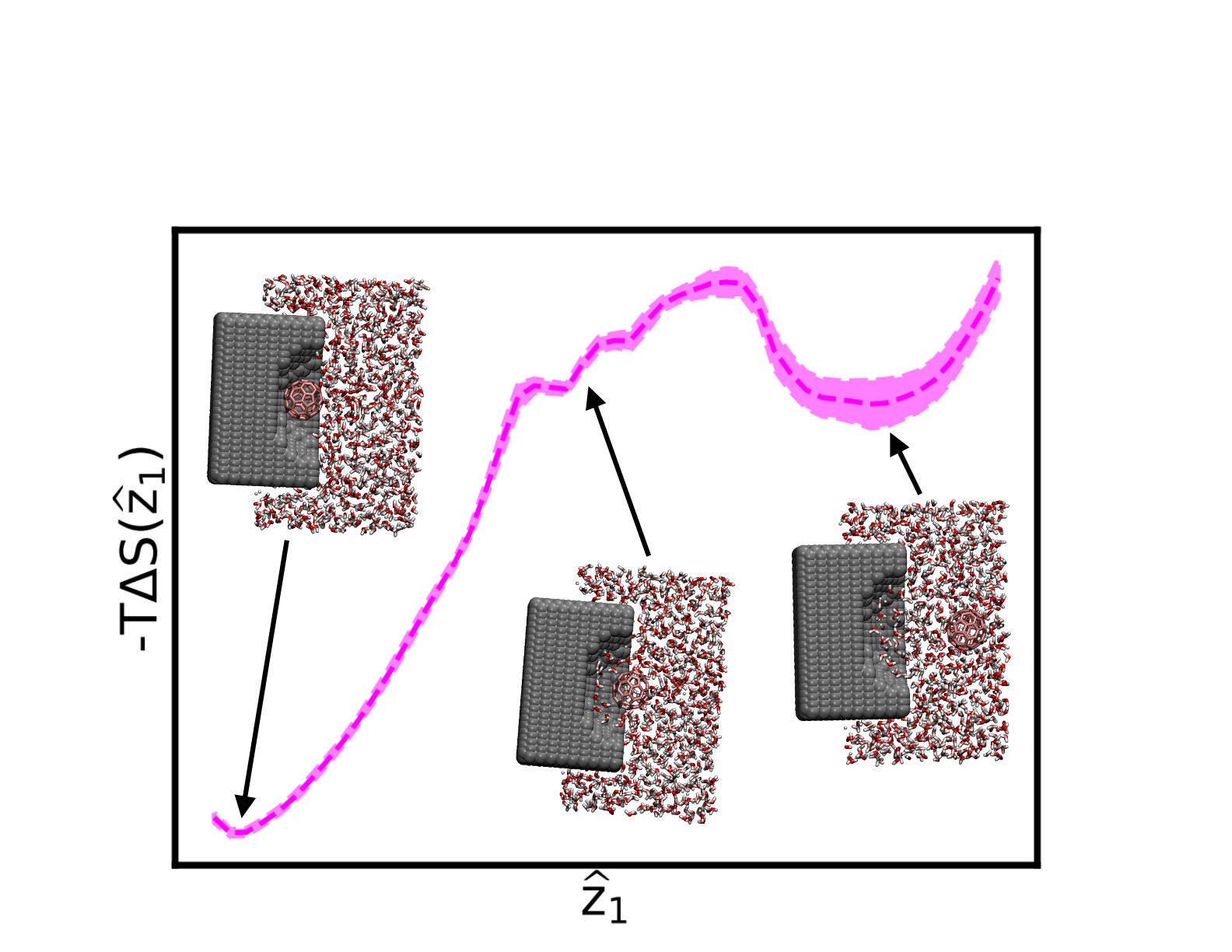
While I studied some nucleation phenomena at UMD, my focus was calculating the thermodynamic profiles of RCs learned using the SPIB approach. One project focused on determining good RCs for the unbinding of two model hydrophobic ligands, a united-atom methane particle and a C60 buckyball fullerene, from rigid, hydrophobic pockets. From this analysis, we confirm that a thermodynamically-optimized, one-dimensional RC for the unbinding for both the methane and buckyball involves surmounting an entropy barrier to unbind, with energetic considerations playing a minor role. Furthermore, we find that solute wetness is the main driver of methane dissociation while pocket wetness plays a major role for the buckyball system until the transition state, after which solute wetness dominates the RC. These observations should yield transferable knowledge to the important features describing ligand unbinding in more complication and physiologically relevant systems, such as drug-protein interactions. This study is published in the Journal of Physical Chemistry B. A preprint is available on the arXiv.